Informal taxonomy to database conversion
In this article we'll convert some tree data into a graph and relational data base.
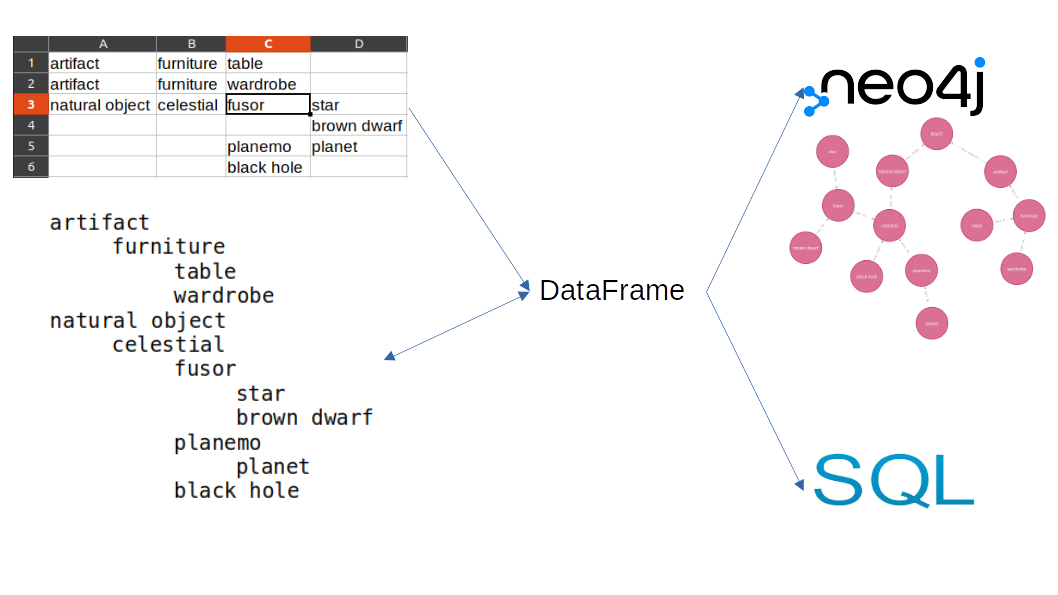
In some cases, we create informal taxonomies from scratch without the aid of sophisticated programs. However, we may need later to formalize and integrate them into a rigurous data base system. Maybe we began using some of the following methods:
| (1) Tabbed text | (2) Mind mapping software | (3) Spread sheet |
|---|---|---|
 |
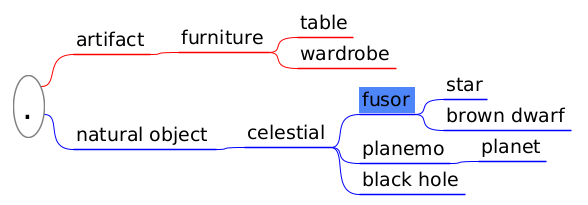 |
 |
Applications like Freeplane allows us to convert its taxonomy into tabbed text and viceversa (by copying and pasting). I will provide a way to convert from tabbed text and spread sheet. Spread sheet can be particulary useful because it allows us to reorder effortlessly the taxonomy following changes, provided that all ancestors cells are filled.
Table to Indexed dataframe
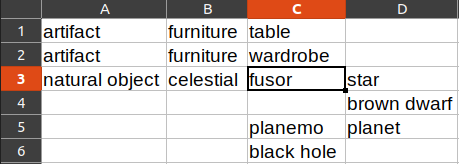
We use this as source data:

We create a python function using pandas
import pandas as pd
def table_to_indexed_dataframe(df):
# remove Nan
df.fillna('', inplace=True)
# remove multiple whitespaces and tabs and newlines
df.replace('\s+', ' ', regex=True, inplace=True)
# strip
df.apply(lambda x: x.str.strip() if isinstance(x, str) else x)
newdata = []
current_values = []
current_parents = []
for index, row in df.iterrows():
lista = row.to_list()
# like rstrip but removing bool(item)=False from the end
while lista and not lista[-1]:
lista.pop()
for pos, text in enumerate(lista):
if pos > len(current_values) - 1: # insertion: parent is always the last one
current_values.append(text)
current_parents.append(len(newdata))
output_row = (len(newdata) + 1, current_parents[-1], str(text), '\t' * pos + str(text))
newdata.append(output_row)
elif text and current_values[pos] != text: # siblings
current_values = current_values[:pos] + [lista[pos]]
current_parents = current_parents[:pos + 1]
parent = current_parents[pos - 1] # previous
output_row = (len(newdata) + 1, current_parents[-1], str(text), '\t' * pos + str(text))
newdata.append(output_row)
newdf = pd.DataFrame(newdata, columns=['id', 'parent_id', 'text', 'tabbed_text'])
newdf = newdf.set_index('id')
return newdf
filename = 'test.tsv'
df = pd.read_csv(filename, sep='\t', header=None)
new_taxonomy_df = table_to_indexed_dataframe(df)
print(new_taxonomy_df)
new_taxonomy_df.to_csv('test_output.csv', sep='\t', index='id')
Result:
parent_id text tabbed_text
id
1 0 artifact artifact
2 1 furniture \tfurniture
3 2 table \t\ttable
4 2 wardrobe \t\twardrobe
5 0 natural object natural object
6 5 celestial \tcelestial
7 6 fusor \t\tfusor
8 7 star \t\t\tstar
9 7 brown dwarf \t\t\tbrown dwarf
10 6 planemo \t\tplanemo
11 10 planet \t\t\tplanet
12 11 terrestrial \t\t\t\tterrestrial
13 6 black hole \t\tblack hole
Tabbed text to Indexed dataframe
def tabbed_text_to_indexed_dataframe(tabbed_text, separator='\t'):
lines = [_ for _ in tabbed_text.splitlines() if _.strip()]
table = [_.split(separator) for _ in lines]
df = pd.DataFrame(table)
return table_to_indexed_dataframe(df)
tabbed_text_sample = """
artifact
furniture
table
wardrobe
natural object
celestial
fusor
star
brown dwarf
planemo
planet
terrestrial
black hole
"""
new_taxonomy_df = tabbed_text_to_indexed_dataframe(tabbed_text_sample)
print(new_taxonomy_df)
Result: same as above
Reverse: Indexed dataframe to tabbed text
def indexed_dataframe_to_tabbed_text(df, parent_id=0, level=0):
rslt_df = df[df['parent_id'] == parent_id]
for index, row in rslt_df.iterrows():
print('\t' * level + row['text'])
indexed_dataframe_to_tabbed_text(df, index, level+1)
Result:
artifact
furniture
table
wardrobe
natural object
celestial
fusor
star
brown dwarf
planemo
planet
terrestrial
black hole
Indexed dataframe to SQL
Create simple creation instructions in SQL thus:
def create_SQL(df):
converted_rows = []
for index, row in df.iterrows():
parent_id, text, tabbed_text = row['parent_id'], row['text'], row['tabbed_text']
converted_rows.append(f"\n\t({index},{parent_id},'{text}','{tabbed_text}')")
sql_expression = f"""CREATE TABLE TAXONOMY
(ID INT PRIMARY KEY
,parent_id INT NOT NULL
,text VARCHAR NOT NULL
,tabbed_text VARCHAR);
INSERT INTO TAXONOMY VALUES{','.join(converted_rows)};
"""
return sql_expression
Result:
CREATE TABLE TAXONOMY
(ID INT PRIMARY KEY
,parent_id INT NOT NULL
,text VARCHAR NOT NULL
,tabbed_text VARCHAR);
INSERT INTO TAXONOMY VALUES
(1,0,'artifact','artifact'),
(2,1,'furniture',' furniture'),
(3,2,'table',' table'),
(4,2,'wardrobe',' wardrobe'),
(5,0,'natural object','natural object'),
(6,5,'celestial',' celestial'),
(7,6,'fusor',' fusor'),
(8,7,'star',' star'),
(9,7,'brown dwarf',' brown dwarf'),
(10,6,'planemo',' planemo'),
(11,10,'planet',' planet'),
(12,11,'terrestrial',' terrestrial'),
(13,6,'black hole',' black hole');
Reverse: SQL to tabbed text
Use this SQL code:
WITH RECURSIVE tree(ID, text, url, level) AS (
SELECT 0, 'ROOT', '', 0
FROM TAXONOMY
UNION
SELECT CHILD.ID, CHILD.text, PARENT.url || '/' || CHILD.ID, PARENT.level + 1
FROM TAXONOMY AS CHILD
JOIN tree AS PARENT ON CHILD.parent_id = PARENT.ID
)
SELECT CONCAT(REPEAT(' ', level), text) FROM tree
ORDER BY url;
Indexed dataframe to graph database (neo4j, using cypher)
Create simple creation instructions in cypher code thus:
def create_graph(df):
exprs = ["CREATE (n%d:Node {name: '%s'})-[:HAS_PARENT]->(n%d)" \
% (index, row['text'], row['parent_id'])
for index, row in df.iterrows() ]
print("CREATE (n0:Node {name: 'ROOT'})\n" + '\n'.join(exprs) + ';')
return "CREATE (n0:Node {name: 'ROOT'})\n" + '\n'.join(exprs) + ';'
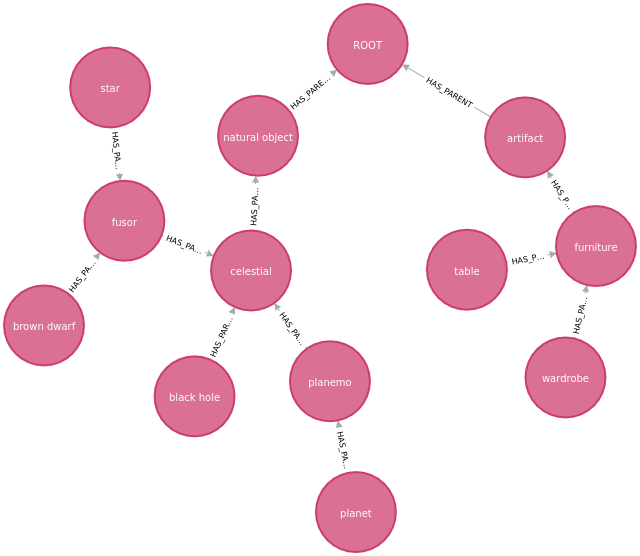
Result: use the cypher code in neo4j browser:
CREATE (n0:Node {name: 'ROOT'})
CREATE (n1:Node {name: 'artifact'})-[:HAS_PARENT]->(n0)
CREATE (n2:Node {name: 'furniture'})-[:HAS_PARENT]->(n1)
CREATE (n3:Node {name: 'table'})-[:HAS_PARENT]->(n2)
CREATE (n4:Node {name: 'wardrobe'})-[:HAS_PARENT]->(n2)
CREATE (n5:Node {name: 'natural object'})-[:HAS_PARENT]->(n0)
CREATE (n6:Node {name: 'celestial'})-[:HAS_PARENT]->(n5)
CREATE (n7:Node {name: 'fusor'})-[:HAS_PARENT]->(n6)
CREATE (n8:Node {name: 'star'})-[:HAS_PARENT]->(n7)
CREATE (n9:Node {name: 'brown dwarf'})-[:HAS_PARENT]->(n7)
CREATE (n10:Node {name: 'planemo'})-[:HAS_PARENT]->(n6)
CREATE (n11:Node {name: 'planet'})-[:HAS_PARENT]->(n10)
CREATE (n12:Node {name: 'black hole'})-[:HAS_PARENT]->(n6);
Consult with
MATCH (n) RETURN n
Erase database with:
MATCH ()-[r]-() DELETE r;
MATCH (n) DELETE n;